Biologists Untangle Growth and Defense in Maize, Define Key Antibiotic Pathways
Researchers identify six genes responsible for the production of plant antibiotics known for critical disease resistance
September 19, 2019
By Mario Aguilera
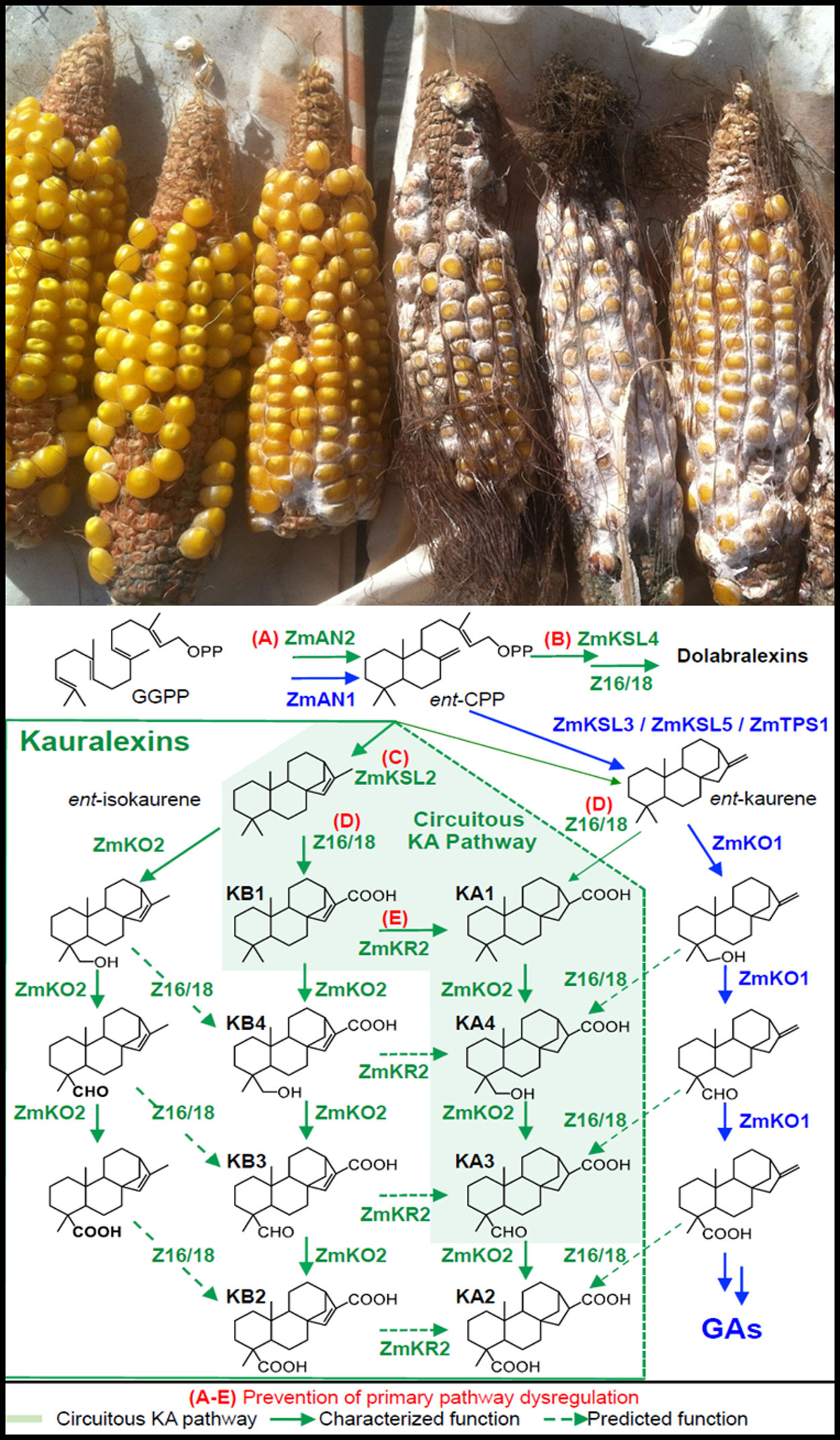
Maize plants are protected from disease-causing fungal pathogens by recent gene duplications in multiple hormone pathways that enable the production of on-demand antibiotics.
In order to meet the demands of growing human populations, agricultural production must double within the next 30 years. Yet the health of today’s crops and the promise of their yield face a rising slate of threats—from pests to chaotic weather events—leading to an urgent need to identify effective, natural plant defense strategies.
Biologists have access to a wealth of genomic and biochemical data, but rapidly deciphering entire biochemical pathways that protect key crops of global significance remains a significant challenge. Scientists are peeling away at the layers of immunity in maize, a staple for diets around the world, to determine if there are key genes that enable surprisingly diverse antibiotic cocktails that can be produced as defensive blends against numerous disease agents.
Now, a systematic and combined approach to identifying such genes in crop defense has been developed by Yezhang Ding, Alisa Huffaker and Eric Schmelz of the University of California San Diego and their colleagues and is described in Nature Plants.
“We need to know which crop defense mechanisms are effective and what we can do sustain or further improve them,” said Schmelz. “Coauthors and collaborators in China are already taking some of the maize genes we characterized and are using them to significantly improve disease resistance in rice.”
Historically, defining a complete new biochemical pathway in crops has required stepwise progress and often the better part of an entire research career. In the new study, the UC San Diego biologists describe how they combined an array of scientific approaches to clearly define six genes that encode enzymes responsible for the production of key maize antibiotics known to control disease resistance.
Maize plants lacking small molecule antibiotic defenses, derived from a skeleton of 20 carbon atoms known as diterpenoids, commonly suffer dramatic increases in fungal disease susceptibility.
“Most people appreciate that pine trees are heavily protected by sticky acid resins that kill or deter a majority of microbes and insects,” said Schmelz. “We describe a complete maize biosynthetic pathway that also makes acid resins on-demand at the site of fungal attack. Interestingly, almost the entire pathway is derived from evolutionarily recent gene duplications from diverse hormone pathways related to plant growth and human testosterone metabolism.”
One of the evolutionary steps was a comparatively recent gene duplication three million years ago from the hormone pathway responsible for plant growth called gibberellins. In a step not clearly borrowed from hormone biosynthesis, two highly promiscuous oxidative enzymes (with broad substrate and product specificity) termed cytochrome P450s were characterized to produce unique reactions different from known conifer pathways. In total, the effort leveraged more than 2,000 plant samples, each with 36,861 transcripts, spanning 300 different maize lines to systematically narrow candidates and define a maize pathway for antibiotics effective against fungal pathogens.
In addition to Ding, Huffaker and Schmelz, coauthors of the paper include Katherine Murphy, graduate students Elly Poretsky and Andrew Sher, along with Evan Saldivar, Mengxi Wu, Zhouxin Shen and Steve Briggs (UC San Diego); Gabriel Castro-Falcon and Chambers Hughes (Scripps Institution of Oceanography, UC San Diego); Sibongile Mafu and Philipp Zerbe (UC Davis); Bing Yang and Si Nian Char (Iowa State University, Ames); Shawn Christensen (Agricultural Research Service, Gainesville, Florida); Qiang Wang (Sichuan Agricultural University); Lexiang Ji and Robert Schmitz (University of Georgia); Karl Kremling and Edward Buckler (Cornell University); and Jorg Bohlmann (University of British Columbia).
The research was funded by the National Science Foundation (NSF) Plant-Biotic Interactions Program (grant 1758976), a Department of Energy Joint Genome Institute Community Science Program (grant CSP2568) and by a fellowship provided by the NSF Graduate Research Fellowship Program. As a graduate student of Huffaker’s lab, Poretsky received partial funding and training under the UC San Diego-NIH training grant.
