New Technologies Depict Transport Machine inside Cells as Having 'Lego-like' Ability
Cryo-EM and cryo-electron tomography among tools used to map previously unseen 3-D features of nuclear pore complex
January 31, 2022
By Mario Aguilera
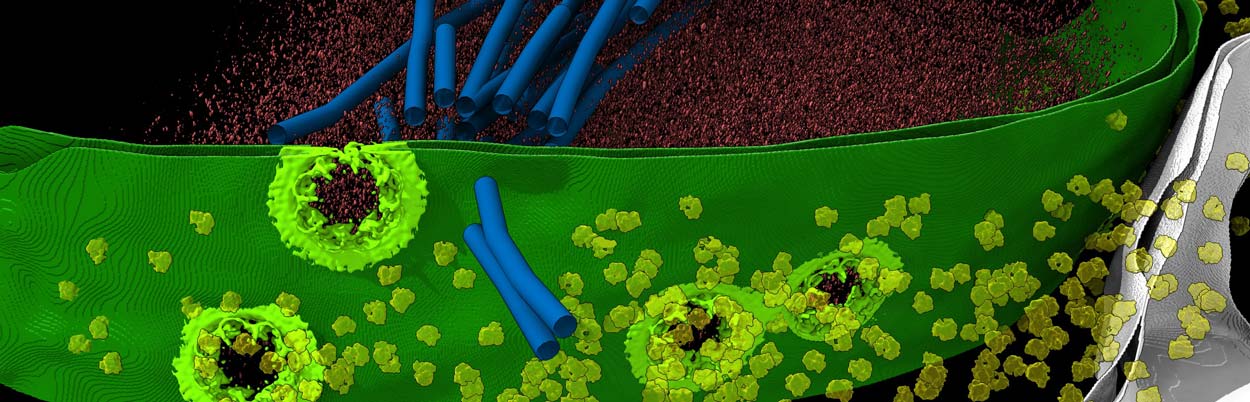
UC San Diego researchers and their colleagues used a variety of advanced techniques to develop the highest-resolution map to date of the nuclear pore complex (seen here in light green), a critical gateway considered the "guardian of the genome."
Like the head of security at a posh nightclub, cells feature a component known as the nuclear pore complex (NPC) that serves as a highly selective gateway allowing molecules to be transported in and out of the nucleus.
Given this critical role as the "guardian of the genome," failure of the NPC to properly function has been linked to many illnesses—including several related to aging, viral infections and certain neurological conditions. The NPC therefore has become the target of new therapeutics to treat such disorders, yet its size, flexibility and dynamic makeup present barriers to unlocking its true structure.
University of California San Diego researchers, including Postdoctoral Scholar Digvijay Singh, Associate Professor Elizabeth Villa and their colleagues at UC San Diego and several institutions, have used new technologies to develop the most comprehensive structural map of the NPC to date. The results were published January 20, 2022 in the journal Cell and offer a new perspective of the cylindrically shaped NPC as a "Lego-" like structure that can have pieces added, deleted or rearranged.
"Critical biology happens at the molecular level, which is also a dominant level for therapeutic strategies like medical drugs. But seeing molecular and sub-molecular features inside cells was long out of reach and just lately made possible," said Singh. "Our new study exemplifies this increasing potential. Our work also illustrates the value of combining different orthogonal methodologies to interpret these molecular level maps into even greater details."
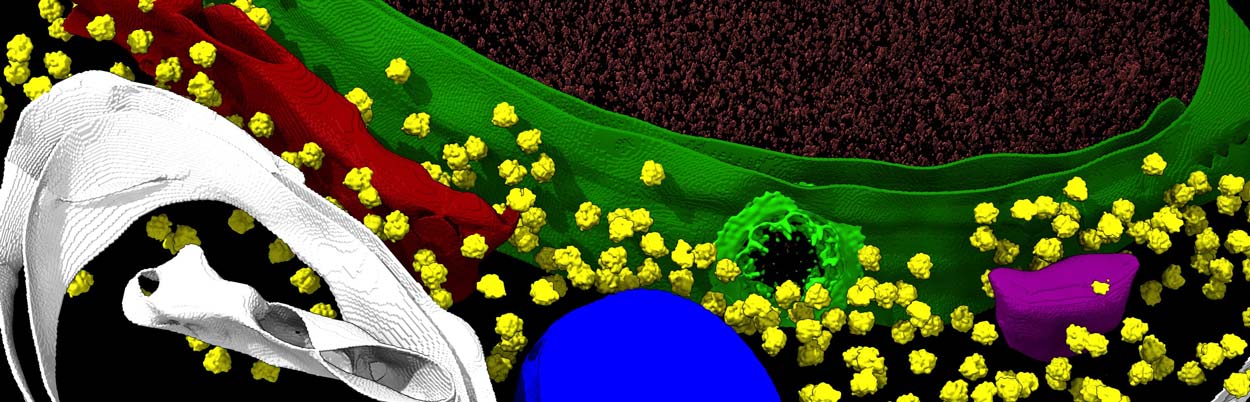
Using cryo-EM, cryo-electron tomography and other techniques, a research team has provided unprecedented insights of the nuclear pore complex, shown in light green against the darker green nuclear membrane.
Using yeast as a model, the researchers employed multiple experimental and computational techniques to develop the new map, including instrumentation featured at UC San Diego’s Cryogenic Electron Microscopy (cryo-EM) facility. The researchers used a focused ion beam to remove ultrathin layers from frozen yeast cells. They then used a transmission electron microscope to obtain two-dimensional images of these thin yeast cell layers from various angles in continued cryogenic conditions. Finally, computational approaches were used to combine those two-dimensional images into a three-dimensional image of the NPC in its natural environment, providing an in situ architecture of the NPC.
Collaborators at Boston and Rockefeller universities isolated NPCs from cells and utilized cryo-EM to obtain the highest-resolution in vitro structure to date. This isolated NPC was more compact than its in situ counterpart. These two structures, one in its functional state inside cells and one obtained through biochemical means, were used as a scaffold where a plethora of data from other techniques was integrated to help the researchers understand: how the NPC moves its parts during the act of transport; the interconnected architecture of its core scaffold; and its connections to the nucleus via a ring of anchors. In addition, different isoforms of NPC were also identified inside the cells for the first time.
"When taken together, the data suggest that at least three NPC isoforms coexist in yeast," the researchers conclude in the paper. "The NPC is a modular assembly with a ‘Lego’-like ability to have major scaffold elements added, reorganized and/or removed."
Other authors in this work included members of the laboratories of Christopher Akey (Boston University); Michael Rout and Javier Fernandez-Martinez (Rockefeller University); Steve Ludtke (Baylor College of Medicine); Andrej Sali (UC San Francisco); and Sue Jespersen (Stowers Institute for Medical Research).
Funding for the research was provided by the Howard Hughes Medical Institute, the National Institutes of Health, the Pew Scholars Program and the Stowers Institute for Medical Research. The Damon Runyon Cancer Foundation supports Singh.
— With information from the Villa Lab
